Metabolic Pathway Prediction of Enzymes for Plant
Based on machine learning approach, this tool is use to predict metabolic pathway of enzymes.
How to cite mApLe:
Almeida RO, Valente GT. Predicting metabolic pathways of plant enzymes without using
sequence similarity: Models from machine learning. The Plant Genome. 2020;e20043. https://doi.org/10.1002/tpg2.20043
More about mApLe: mApLe: Predicting metabolic pathways of enzymes.
Tutorial
To use mAppLe tool, you will need:
- OS Linux (Ubuntu 18.04 or higher)
- R 3.6.0 or higher
- Memory RAM: 8GB or higher
- MultiCore processor
Download:
mApLe.tar.xz - Uncompress in /home/USER
mApLe - Reference Manual - How to use mApLe
Raw Data - Data used to create the models
All Dataset - Dataset used to training, test and validation
mApLe-test subset 1 - Dataset used in mApLe, DeepEC and KAAS
About mAppLe
You can visualize the prediction table, plot and a summary of data in mAppLe tool.
You can download (as compressed file) the results: - Table of Results; - Weka files results; - Plots; - ARFF files used; - Attributes generated.
Table of results will show like the follow figure:
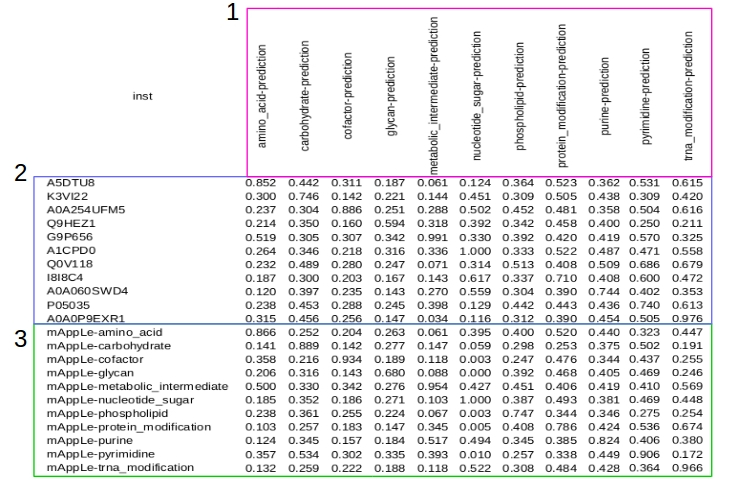
1 - Metabolic pathways analysed.
2 - Identification of sequences and probability results.
3 - Sequence control of mApLe tool and probability results.